Submission Guidelines
Ground rules for submission
- The quality of the information depends solely on submitters.
- Any model can be shared to the public if fulfilling minimum requirements (described in the next steps).
- If a model refers to a publication, it has to have the same or equivalent mathematical representation. If this is not the case, the publication can still be referred to but modifications to the publication and their motivations needs to be clearly detailed in the documents uploaded.
- Uploading of observed human data is not accepted. This might be changed in the future as the project evolves.
-
A simulated data set is needed to let other users:
- understand how the data set has to be constructed, i.e. which data items, specific records, etc., are needed;
- run the model, e.g. execute estimation/simulation OR execute an evaluation (MAX=0 in NONMEM): the type of execution is the submitter's decision.
Therefore, a data set with the dependent variable simulated from the uploaded model OR with a dummy dependent variable is acceptable.
-
Output files are needed to
- prove the model is executable (output from execution of simulated data is sufficient for this purpose);
- report the final model parameter estimates and corresponding uncertainty (output from execution of real data is needed for this purpose).
When real data are accessible to the submitter, the output file obtained from the real data needs to be provided. When real data are not available, an output file containing the information about parameter estimates and their uncertainty, as available in the publication, needs to be provided.
Create relevant login credentials
If you do not have an account on the DDMoRe Repository, please register and you will be sent a password that will allow you to log on and start the submission process. Your name will be displayed as submitter for the models you upload, thus define it appropriately: first name, space, last name (you can also modify it once logged in by selecting `Edit User' from your profile page).
Identify the scenario
To allow as many models as possible to be uploaded, a number of submission scenarios have been identified based on the possibility to generate MDL and/or PharmML code. The documentation required to upload a model varies based on the scenario chosen. Therefore, please use the decision tree below to identify which case is applicable to your model. For scenarios 1, 2 and 4, example models are given below, which may be helpful.Once that is done, follow carefully the instructions below for the chosen scenario, all steps are mandatory if not differently specified.
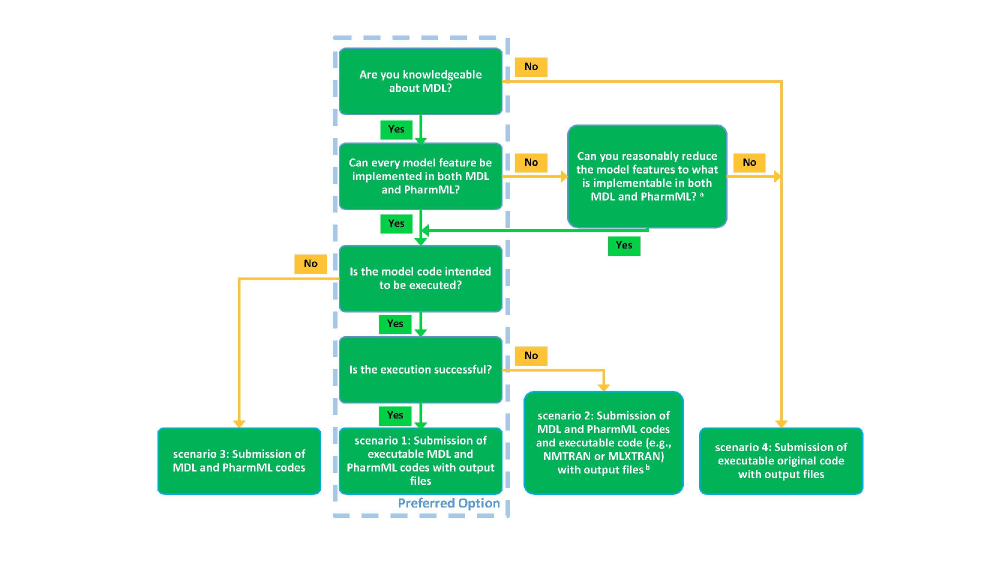
In the text for each scenario below, the minimal requirements for publication are underlined.
Follow the file naming rules
Before preparing the material to be uploaded to the DDMoRe Repository, please look at the naming rules below, covering the files to be uploaded below.
Go to scenario 1 | scenario 2 | scenario 3 | scenario 4
Scenario 1: Executable MDL AND PharmML code
This applied when the original model can be successfully coded in both MDL and PharmML and that the MDL and PharmML code can be executed in the Interoperability Framework.
Example model: Friberg_2009_Schizophrenia_Asenapine_PANSS [http://repository.ddmore.foundation/model/DDMODEL00000002]
- Start submission by uploading PharmML code (version up to 0.6.1) as the mandatory file (the DDMoRe Repository will thus provide an automatic human-friendly model visualization within the Model Definition tab).
-
Upload the following additional files. Add a short and meaningful description in the box next to each file.
- MDL code (version 7.0 or later supported by DDMoRe Interoperability Framework 4.1 or later).
- Real data (possible if non-human) or simulated data. If simulated: provide a dataset similar to the real dataset with dependent variable(s) simulated from the uploaded model OR with dummy dependent variable(s), and with covariates replaced by their typical values.
- Command.txt, which describes the successful execution command/script: this file also needs to specify the uploaded file names (for input data, executable model and output results of the execution) and the used language/product versions/target tool, see example 1.
- Output file obtained after execution of uploaded dataset and MDL model code.
- Additional output file (if you decided to upload simulated data), with the final model parameter estimates and the corresponding uncertainty. If you have access to real data and do not upload them, this is the output file obtained after execution with real data. If you do not have access to real data but the model refers to a publication, this file should include final estimates and corresponding uncertainty as given in the publication used as reference, see output_example.
-
Model_Accommodations.txt if you had to modify the published model: describe in this file the differences between the implemented model and the model in the publication used as reference. Use this template and see also examples in the following two model entries:
Hamren_2008_diabetes_tesaglitazar [http://repository.ddmore.foundation/model/DDMODEL00000003],
Trefz_2015_metabolism_Kuvan_TurnoverKPD [http://repository.ddmore.foundation/model/DDMODEL00000103]. - Any other files considered to be useful.
-
After pressing Upload, fill in Model Information.
In the box Name: change the default to a describion of your model (max 150 characters). This is the descriptive name which will be shown when browsing the DDMoRe Model Repository (see [http://repository.ddmore.foundation/models] for examples). - In the box Description: add a brief and meaningful description of your model. This text is shown in the Overview tab, but not directly in the browser of the DDMoRe Model Repository.
- Next add a reference for your model. If the model is related to a publication, provide the corresponding publication identifier: PubMed ID (preferred), DOI, other link (URL) or choose "Publication without link".
- Before pressing "Complete Submission", check the "Summary of your submission" (click on "Back" within the repository and refine if necessary).
- After completing the submission, enter the model annotations by pressing the button "Annotate" (see buttons on the left-had bar) and fill in the fields on all available tabs, thenpress "Save" to save the information you provided and "Validate" to check the validity of the annotations you entered.
- Click on "Return to model display page" and then on the "Publish" button (see the buttons on the left-hand bar) in order to share your model with the community and complete the publishing process. Before publishing, please ensure you provided all the files and information described in previous steps, otherwise the (automatic) publishing process will reject the model publication. If you need to modify your model to have it published, click on the button "Update" on the left-hand bar.
Scenario 2: MDL and PharmML codes and executable code
This scenario applies when the MDL and PharmML code cannot be entirely executed in the DDMoRe Interoperability Framework (via translation of MDL and PharmML to the target tool language) and an additional executable code (e.g. NMTRAN, MLXTRAN) must be provided.
Example model: Hamren_2008_diabetes_tesaglitazar [http://repository.ddmore.foundation/model/DDMODEL00000003]
- Start submission by uploading a non-executable PharmML code (version up to 0.6.1) as mandatory file (the Model Repository will thus provide an automatic human-friendly model visualization within the Model Definition tab).
-
Upload the following as additional files. Add a short and meaningful description in the box next to each file.
- A non-executable MDL code (version 7.0 or later supported by DDMoRe Interoperability Framework 4.1).
- An executable code (e.g. NMTRAN, MLXTRAN). The submitter is encouraged to use the MDL or PharmML code, convert it into the target tool language usingthe DDMoRe Interoperability Framework, and modify the produced code in order to make it executable.
- Real data (possible if non-human) or simulated data. If simulated: provide a dataset similar to the real dataset with dependent variable(s) simulated from the uploaded model OR with dummy dependent variable(s), and with covariates replaced by their typical values.
- Command.txt, which describes the successful execution command/script: this file also needs to specify the uploaded file names (for input data, executable model, and output results of the execution) and the used language/product versions/target tool. Moreover, the modifications mentioned in point b) above need to be described within this file: as an example, see Hamren_2008_diabetes_tesaglitazar [http://repository.ddmore.foundation/model/DDMODEL00000003].
- Output file obtained after execution of uploaded dataset and executable code.
- Additional output file if you decided to upload simulated data, with the final model parameter estimates and the corresponding uncertainty. If you have access to real data this is the output file obtained after execution with real data. If you do not have access to real data and do not upload them, this file should include final estimates and corresponding uncertainty as given in the publication used as reference, see output_example.
-
Model_Accommodations.txt if you had to modify the published model: describe in this file the differences between the implemented model and the model in the publication used as reference (any changes that were required in the executable code to make it run should not be stated here but in the Command.txt file). Use this template and see also examples in the following two model entries:
Hamren_2008_diabetes_tesaglitazar[http://repository.ddmore.foundation/model/DDMODEL00000003] and
Trefz_2015_metabolism_Kuvan_TurnoverKPD [http://repository.ddmore.foundation/model/DDMODEL00000103]. - Any other files considered to be useful.
-
After pressing Upload, fill in Model Information.
In the box Name: change the default to a description of your model (max 150 characters). This text is the descriptive name which will be shown when browsing the DDMoRe Model Repository (see http://repository.ddmore.foundation/models for examples). - In the box Description: add a brief and meaningful description of your model. This text is shown in the Overview tab, but not directly in the browser of the DDMoRe Model Repository. Next add a citation for your model. If the model is related to a publication, provide the corresponding publication identifier: PubMed ID (to be preferred), DOI, other link(URL) or choose "Publication without link".
- Before pressing "Complete Submission", check the "Summary of your submission" (click on "Back" within the DDMoRe Model Repository and refine if necessary).
- After completing the submission, enter the model annotations by pressing the button "Annotate" (see the buttons on the left-hand bar) and fill in the fields on the all available tabs; then press "Save" to save the information you provided and "Validate" to check the validity of the annotations you entered. If the annotations are correct a message turns up on top stating ‘Annotations are correct’.
- Click on "return to model display page" and then click on the "Publish" button (see buttons on the left-hand bar) in order to share your model with the community and complete the publishing process. Before publishing, please ensure you provided all the files and information described in the previous steps otherwise the (automatic) publishing process will reject the publication of the model. If you need to modify your model to have it published, click on the "Update" button in the left-hand bar.
Scenario 3: MDL and PharmML codes, not intended for execution
This scenario applies when the model is not intended to be executed in the Interoperability Framework.
Example model: None currently
- Start submission by uploading a non-executable PharmML code (version up to 0.8.1) as mandatory file (the DDMoRe Model Repository will thus provide an automatic human-friendly model visualization within the Model Definition tab).
-
Upload the following additional files. Add a short and meaningful description in the box next to each file.
- MDL code (version 7.0 or later supported by DDMoRe Interoperability Framework 4.1 or later).
-
Model_Accommodations.txt if you had to modify the published model: describe in this file the differences between the implemented model and the model in the publication used as reference. Use this template and see also examples in the following two model entries:
Hamren_2008_diabetes_tesaglitazar [http://repository.ddmore.foundation/model/DDMODEL00000003] and
Trefz_2015_metabolism_Kuvan_TurnoverKPD [http://repository.ddmore.foundation/model/DDMODEL00000103]. - Any other files considered to be useful.
-
After pressing Upload, fill in Model Information.
In the box Name: change the default to a description of your model (max 150 characters). This is the name which will be shown when browsing the DDMoRe Model Repository (see http://repository.ddmore.foundation/models for examples).
In the box Description: add a brief and meaningful description of your model. This text is shown in the Overview tab, but not directly in the browser of the DDMoRe Model Repository. - Next add a reference for your model. If the model is related to a publication, provide the corresponding publication identifier: PubMed ID (to be preferred), DOI, other link(URL) or choose "Publication without link".
- Before pressing "Complete Submission", check the "Summary of your submission" (click on "Back" within the DDMoRe Model Repository and refine if necessary).
- After completing the submission, enter the model annotations by pressing the button "Annotate" (see the buttons on the left-hand bar) and fill in the fields on the all available tabs; then press "Save" to save the information you provided and "Validate" to check the validity of the annotations you entered. If annotations are correct a message turns up on top stating ‘Annotations are correct’.
- Click on "Return to model display page" and then on the "Publish" button (see the buttons on the left-hand bar) in order to share your model with the community and complete the publishing process. Before publishing, please ensure you provided all the files and information described in the previous steps, otherwise the (automatic) publishing process will reject the model publication. If you need to modify your model to have it published, click on the "Update" button in the left-hand bar.
Scenario 4: Executable original code
This scenario applies when the model submitter wants to publish a model that for various reasons has not been coded in MDL and PharmML.
Example model: CMS_Colistin_PK_Karaiskos_2015 [http://repository.ddmore.foundation/model/DDMODEL00000130]
- Start submission by uploading executable original code as the mandatory file (thus, the DDMoRe Model Repository will NOT be able to provide an automatic human-friendly visualisation).
-
Upload the following as additional files. Add a short and meaningful description in the box next to each file.
- Real data (possible if non-human) or simulated data (if simulated: provide a dataset similar to the real dataset with dependent variable(s) simulated from the uploaded model OR with dummy dependent variable(s), and with covariates replaced by their typical values.
- Command.txt, which describes the successful execution command/script: this file also needs to specify the uploaded file names (for input data, executable model, and output results of the execution) and the used target tool, see example 2.
- Output file obtained after execution of uploaded dataset and executable original model code (it is possible to use equivalent to MAX=0 from NONMEM to save time if simulated dataset is used).
- Additional output file if you decided to upload simulated data, with the final model parameter estimates and the corresponding uncertainty. If you have access to real data this is the output file(s) obtained after execution with real data. If you do not have access to real data, this file should include final estimates and corresponding uncertainty as given in the publication used as reference, see output_example.
-
Model_Accommodations.txt if you had to modify the published model: describe in this file the differences between the implemented model and the model in the publication used as reference. Use this template and see also examples in the following two model entries:
Hamren_2008_diabetes_tesaglitazar [http://repository.ddmore.foundation/model/DDMODEL00000003],
Trefz_2015_metabolism_Kuvan_TurnoverKPD [http://repository.ddmore.foundation/model/DDMODEL00000103]. - Any other files considered to be useful
-
After pressing Upload, fill in Model Information.
In the box Name: change the default to a description of your model (max 150 characters). This is the descriptive name which will be shown when browsing the repository (see http://repository.ddmore.foundation/models for examples).
In the box Description: add a brief and meaningful description of your model. This text is shown in the Overview tab, but not directly in the browser of the DDMoRe Model Repository. - Next add a reference for your model. If the model is related to a publication, provide the corresponding publication identifier: PubMed ID(to be preferred), DOI, other link(URL) or choose "Publication without link".
- Before pressing "Complete Submission", check the "Summary of your submission" (click on "Back" within the DDMoRe Model Repository and refine if necessary).
- After completing the submission, enter the model annotations by pressing the button "Annotate" (See buttons on the left-hand bar) and fill in the fields on the all available tabs; then press "Save" to save the information you provided and "Validate" to check the validity of the annotations you entered. If annotations are correct a message turns up on top stating ‘Annotations are correct’.
- Click on "Return to model display page" and then on "Publish" (see the buttons on the left-hand bar) in order to share your model with the community and to complete the publishing process. Before publishing, please ensure you provided all the files and information described in the previous steps, otherwise the (automatic) publishing process will not publish your model. If you need to modify your model to have it published, click on the"Update" button on the left-hand bar.
Naming rules for all uploaded files
-
Data
- Real_xxx — real, non-human
- Simulated_xxx — simulated
-
Executable model code
- Executable_xxx — the model code executable. The executable file is a file which can be executed. With scenario 1, at least two files are executable: the MDL and the PharmML codes. With scenario 2, this name cannot be used for the MDL and PharmML codes. Make sure that the name of the input data in the model code is in agreement with the data file you upload.
-
Non-executable model code
- All files which are not executable must not start with Executable_xxx. In particular: for scenarios 2, and 3, model codes written in MDL or PharmML cannot be named Executable_xxx.
-
Execution command/script
- Command.txt Make sure that the names of the input data, executable code and output file in the script are in agreement with the files you upload.
-
Output file
- Output_real_xxx — from execution using realdata. If real data are not uploaded, this is the additional output file introduced above, i.e. a file with the final model parameter estimates and the corresponding uncertainty obtained either from publication or from execution with real data.
- Output_simulated_xxx — from execution usingsimulated data. Make sure that, if shown, the names of the input data and executable code in this file are in agreement with the files you upload.
-
Modifications with respect to the published model
- Model_Accomodations.txt — text file describing how the mandatory uploaded model differs from the related publication referred to.
Go to scenario 1 | scenario 2 | scenario 3 | scenario 4
File content examples
Command.txt — example 1
Follow this example for the mandatory Command.txt file if scenario 1 aplies. It is highly recommended to follow this example with the initial block of commented lines filled in.
###### Scenario = 1
###### MDL version = <MDL version, constrained to be 7.0 at the moment >
###### IO product = <interoperability product number, constrained to be 4.1 at the moment >
###### Input data = <file name of uploaded input data >
###### Executable model = <file name of uploaded executable >
###### Output = <file name of uploaded output from executable >
#Initialisation
rm(list=ls(all=F)) #clean your workspace first
#Set working directory
myfolder <- "" # name of your project folder
setwd(file.path(Sys.getenv("MDLIDE_WORKSPACE_HOME"),myfolder))
#Set name of .mdl file and dataset for future tasks
mymodel <- ""
datafile <- ""
mdlfile <- paste0(mymodel,".mdl")
#ESTIMATE model parameters using Monolix
mlx <- estimate(mdlfile, target="MONOLIX", subfolder="Monolix")
# Print the estimated parameters
parameters_mlx <- getPopulationParameters(mlx, what="estimates")$MLE
print(mlx)
# Perform model diagnostics for the base model using Xpose functions
mlx.xpdb <- as.xpdb(mlx,datafile)
pdf("GOF_MLX.pdf")
print(basic.gof(mlx.xpdb))
print(ind.plots(mlx.xpdb))
dev.off()
# This code can be expanded upon users needs to e.g. include execution in other tools
Go to scenario 1 | scenario 2 | scenario 3 | scenario 4
Command.txt — example 2
Follow this example for the mandatory Command.txt file if scenario 4 applies. In this specific case, the original code is run with NONMEM via PsN.It is highly recommended to follow this example with the initial block of commented lines filled in. Additional information can be added as required and/or desired.
###### Scenario = 4 ###### PsN version = <PsN version> ###### Original tool version = <NONMEM version> ###### Input data = <file name of uploaded input data> ###### Executable model = <file name of uploaded executable> ###### Output = <file name of uploaded output from executable> execute run1.mod
Go to scenario 1 | scenario 2 |scenario 3 | scenario 4
Output_real
This is an example for the Output_real_xxx file, to be used when the submitter has compiled with the rules of the Repository and has refrained from uploading real human data. It can be a text file or a pdf file with a table, showing the final model parameter estimates and the corresponding uncertainty as provided in the related publication. The text file can be arranged as follows,or it can be an excerpt of a table from the publication; however make sure that notation of parameters is consistent with the uploaded model file (if not add explanation in this file):
|
Parameter name [as in the model] |
Parameter estimate (unit) |
Relative standard error (%) [or Standard error; as given in publication] |
| CL | 104.5 (L/h) | 15% |
| V | 25.4 (L) | 23% |
| etc. |
Go to scenario 1 | scenario 2 |scenario 3 | scenario 4
Model_Accommodations.txt — example
This file has to be uploaded when the model is not implemented as in the original publication.
###### Deviations from published model: ###### Motivation to deviations:
Go to scenario 1 | scenario 2 |scenario 3 | scenario 4